RxnGPT2: Reaction-prompted Sequence Generator
Background
The goal of enzyme engineering in catalytic reaction space is searching for the most potent enzyme variant that efficiently catalyzes a reaction of interest. A folded enzyme sequence accommodates a substrate (chemical compound) binding to a catalytic site and converts it to a product (a beneficial form of chemical compound via natural mechanism as an alternative to chemical synthesis).
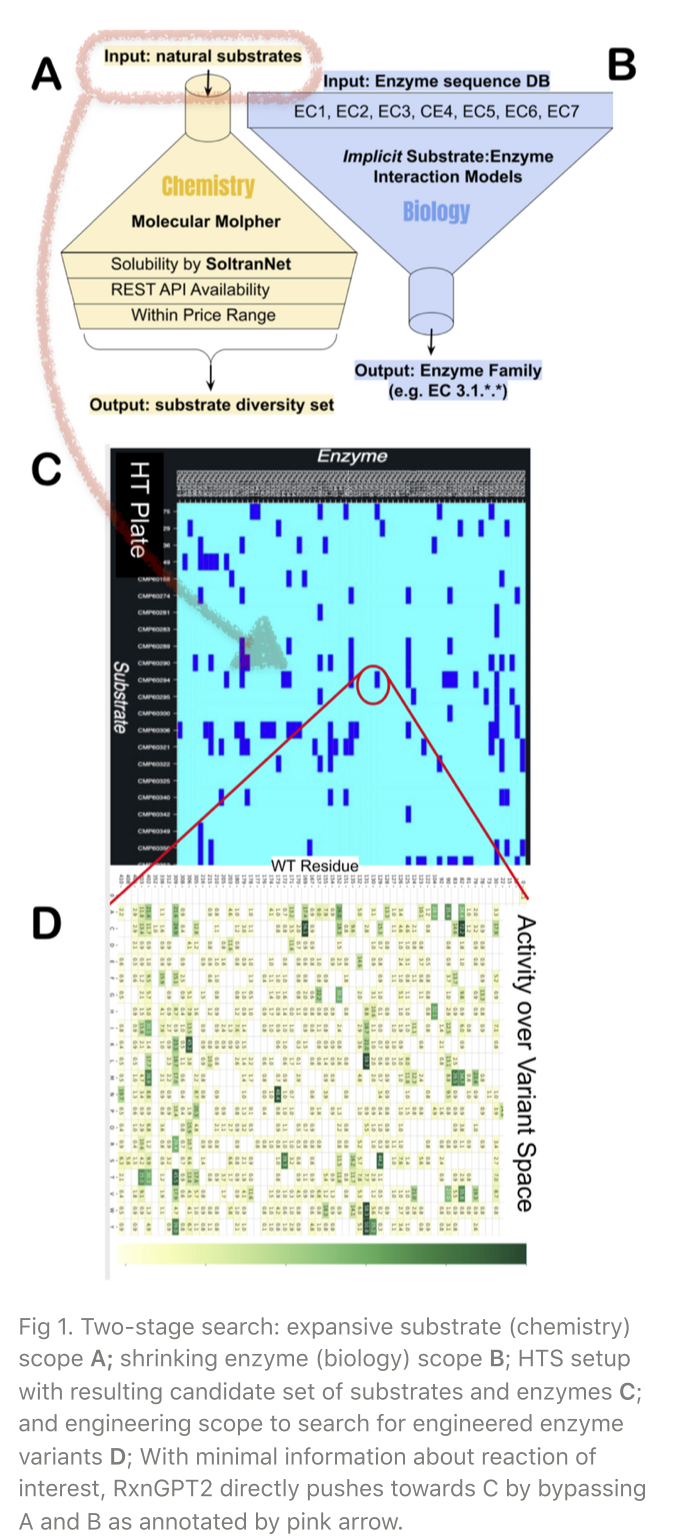
To initiate an engineering scope to search for engineered enzyme variants shown in Fig 1D, we should first find a hit pair of indexed (wild-type) enzymes and substrates from the high-throughput screening (HTS) shown in Fig 1C.
A traditional approach to setting up an HTS is a separate, two-stage search: (1) expansive substrate (chemistry) scope and (2) shrinking enzyme (biology) scope, as depicted in Fig 1A-B. A plate of HTS is filled up with the resulting substrates and enzymes candidate set. Due to uncoupled searches, however, this traditional approach not only takes time but also undergoes off-target problems.
RxnGPT2
To accelerate a two-stage search, more importantly, to ensure tight coupling between substrate and enzyme scoping, we build a reaction-prompted, enzyme sequence generator large language model, RxnGPT2, via three steps: (1) generalizing reaction space; (2) constructing substrate-enzyme knowledge association data; and (3) end-to-end approach from a reaction of interest all the way to catalytic site elucidation.
(1) Generalizing reaction space
Reaction similarity is different from chemical structure similarity. Semantic similarity in reaction space can be attained by utilizing atomic mapping from reactant side onto product side [1]. Subsequently we identify the atoms undergoing a change within a various radius [2]. Consequently, we can search similar reactions semantically, instead of traditional chemical structure (such as SMILES strings) or fingerprint pattern-based search.
(2) Constructing substrate-enzyme knowledge association data
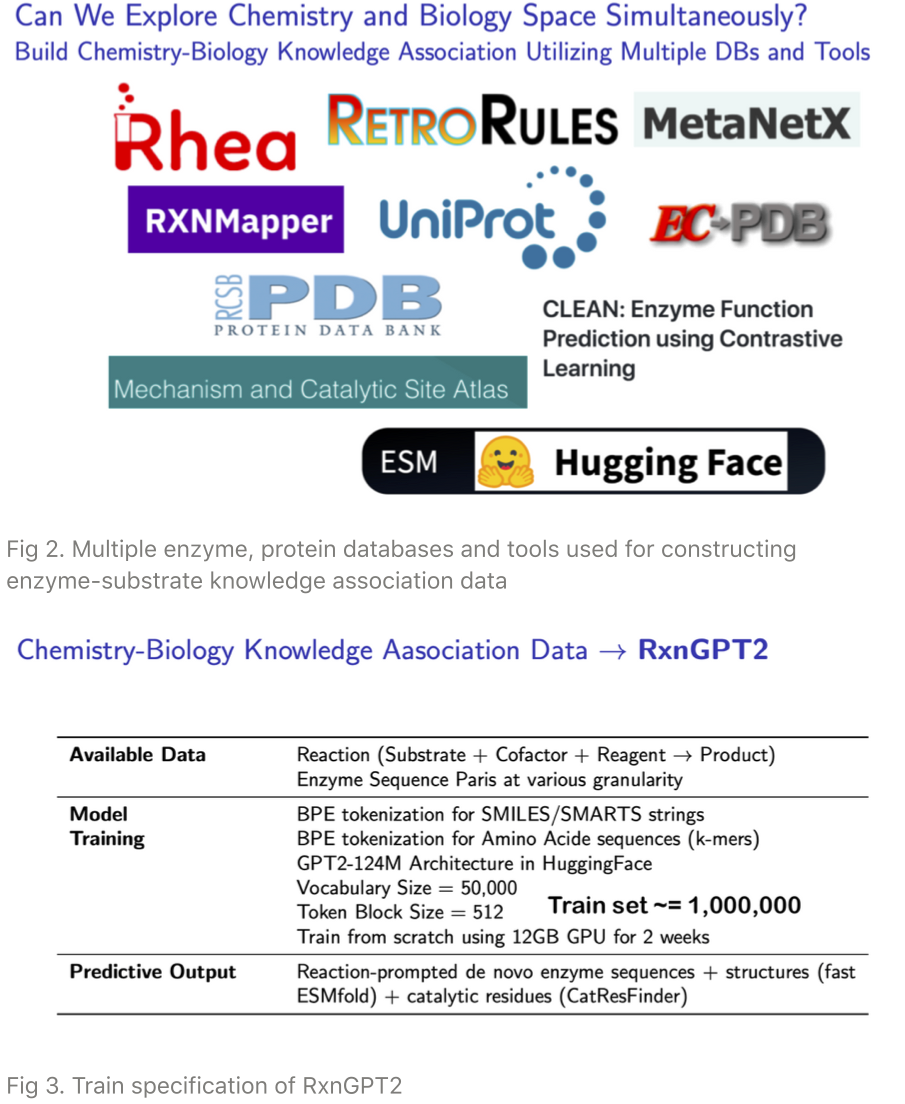
With the generalized reaction data, we then associate a reaction to corresponding enzyme sequence by utilizing multiple databases such as Rhea and UniProt (comprehensive database and tools listed in Fig 2).
(3) End-to-end approach from a reaction of interest all the way to catalytic site elucidation.
With the coupled data as our train set, we build a RxnGPT2 model by
training from scratch with a base architecture of GPT2-124M . Our input
data consists of {prompt}<sep>{response},
where {prompt} is a
generalized catalytic reaction at various granularities, and {response} is an
enzyme sequence. We used Byte-Pair-Encoding (BPE) for both chemical
reaction strings and amino acid sequence strings. This is different from 20
amino acid based tokenization used in other protein language models.
Moreover, to our knowledge, there is no such generative model yet
tokenizing both chemical language and protein language simultaneously.
Detailed training specification in terms of trainset, tokenization, and GPU
usage is summarized in Fig 3.
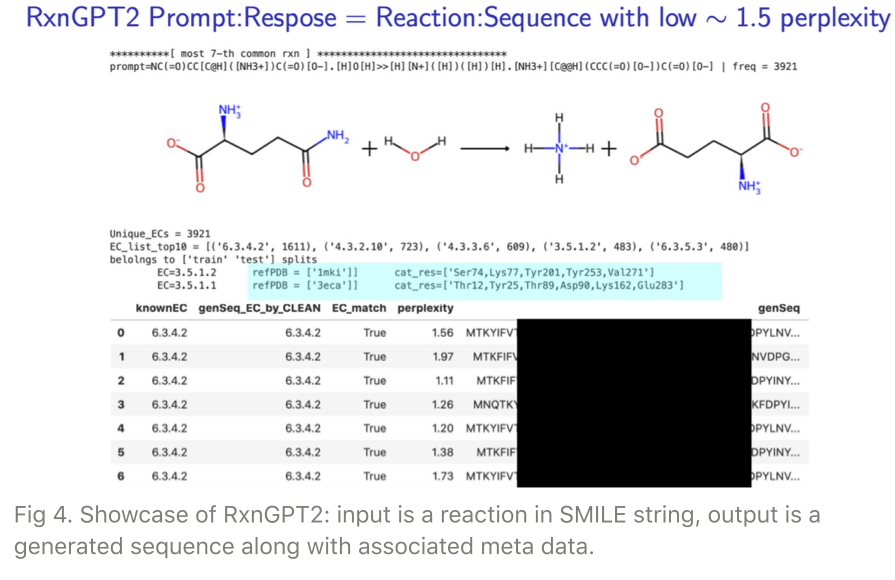
Fig 4 demonstrates a showcase of the RxnGPT2: reaction-prompted, generated sequences in tabular form with extra information especially a low perplexity (metric of quality of generated sequences).
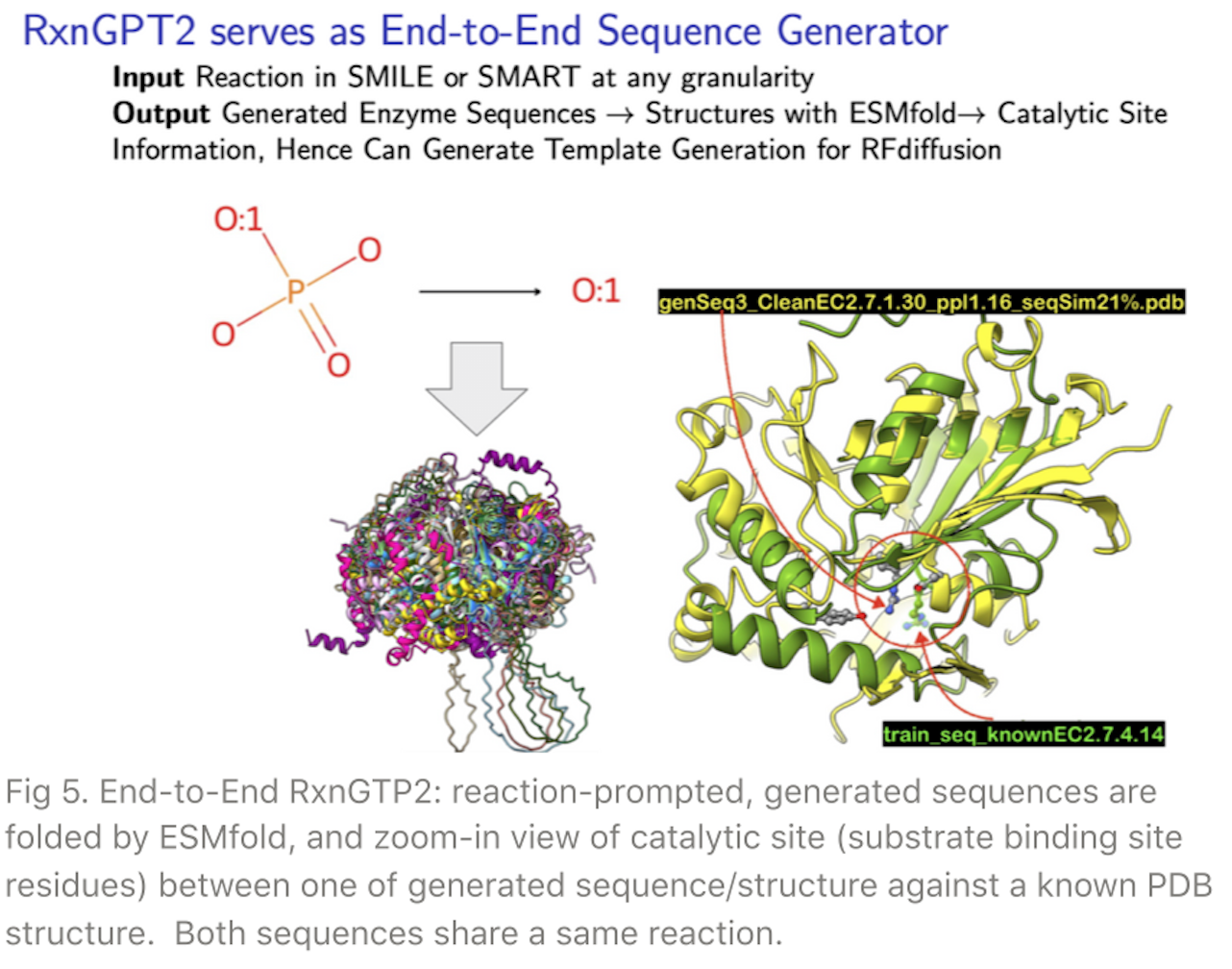
A truly end-to-end application with the RxnGPT2 is exhibited in Fig 5. Users just enter a reaction of interest to the generator at any granularity either in SMILES or SMARTS, and the RxnGPT2 returns an elucidated catalytic site view on the folded structure from the generated sequence.
In conclusion, the RxnGPT2 bypasses traditional two-stage substrates and enzyme scopes, directly pushes forward with tightly coupled, End-to-End, one-stop approach without extensive prior knowledge on the catalytic site structural information. It is a promising generative AI for enzyme discovery.
References
[1] Schwaller P, Hoover B, Reymond JL, Strobelt H, Laino T. Extraction of organic chemistry grammar from unsupervised learning of chemical reactions. Sci Adv. 2021 Apr 7;7(15):eabe4166. doi: 10.1126/sciadv.abe4166. PMID: 33827815; PMCID: PMC8026122.
[2] Kannas C, Thakkar A, Bjerrum E, Genheden S. rxnutils – A Cheminformatics Python Library for Manipulating Chemical Reaction Data. ChemRxiv. Cambridge: Cambridge Open Engage; 2022; This content is a preprint and has not been peer-reviewed.